Blog by 4D-Reef ESRs Daniel Christoph Schürholz, Dino Ramos and Estradivari, based at respectively the Max-Planck-Institute for Marine Microbiology, University of Granada and Leibniz-zentrum fur Tropical Marine Research.
Digitizing the Spermonde coral reefs: a paradise of corals, turf and algae
Finally! During August of last year (2022), we went to our promised field sites in the beautiful sandy islands of the Spermonde Archipelago, just in front of the bustling city of Makassar – South Sulawesi – Indonesia. This archipelago is located in the heart of the marine biodiversity hotspot known as the Coral Triangle, a dream destination for marine researchers. Almost three years we waited to go, and just after the pandemic regulations and global crises relaxed a tiny bit, we packed our (many) bags and went to the field. Each one of us early stage researchers (ESRs) had planned their sampling strategies and locations carefully, considering our research goals, but also accounting for possible collaborations within the team and/or with our amazing partners from the local Hasanuddin University (UNHAS) in Makassar. This specific story is about the collaboration of three 4D-REEF ESRs: Estra, Dino and Daniel, with three outstanding Indonesian students from UNHAS: Phita, Gunawan and Agung, who all flew from the warm Indonesian weather to the frosty German winter.

The digitizers working hard at the Leibniz Centre for Tropical Marine Research ZMT (from left to right): Estra, Gunawan, Agung, Dino, Phita, Daniel.
Something in common: reef benthos
To collaborate within our small team we had to identify what binds our three research projects together. And luckily, the link was easy to find, since in all our cases we are interested in the coral reef benthos (a.k.a., the habitat on the seafloor).
In Estra’s project, for example, she is looking at the ecological implications of shifts from hard corals to turf algae (assemblages of many diminutive species of algae) in turbid reefs. Turf algae and corals live in balance unless the ecosystem is disturbed (usually by man-made pressures), in which case the loss of coral could mean an overexpansion of turf algae. Estra, along with four students from UNHAS, studies the reef’s turf algae mats and how they change over time, as well as their role in the ecosystem (i.e., trapping sediments and detritus – Gunawan; providing nutritious food for herbivorous fish – Munawarrah; competing of space with hard corals and other biotas – Agung) and their interactions with other organisms, such as corals and fish (Puspita).

Turf algae mats covered dead corals on Samalona Island. In reefs near Makassar City (e.g., Lae Lae, Samalona), where sedimentation and river run-off were significant, turf algae mats can cover more than 50% of the bottom substrate and trap a large amount of sediment and detritus. Offshore reefs exhibit fewer turf algae assemblages and sediments/detritus trapped within turf algae (e.g., Kapoposang, Karang Kassi).
Dino’s research focuses on the crustose coralline algae (CCA), a type of red algae with calcified thalli (term referring to algal bodies). They are a key player in the ecosystem, as they are considered to literally build the reef framework together with stony corals (juvenile corals also seem to prefer them when selecting a place to settle). Dino is looking into the diversity of these often overlooked organisms and how their communities shift with varying conditions in the reef, particularly turbidity, in the Spermonde Archipelago.

While not immediately noticeable in most coral reefs, you only need to look in the spaces among the corals to find the ubiquitous CCA. Like the more colorful corals, they can take various morphologies such as the branching form seen in the right image in between the Acropora. Currently twenty species of CCA have been described in the Spermonde Archipelago based on morpho-anatomical characters. This is likely an underestimation based on the abundance of cryptic species detected in recent studies of CCA using DNA. With a lack of basic taxonomic knowledge, even less is known about each of these species’ ecological functions!
Daniel’s project targets the creation of maps of the coral reefs, describing the whole benthic ecosystem by applying advanced underwater imaging techniques and artificial intelligence (AI) pipelines. Understanding the spatial distribution of organisms on the maps can help to identify their specific life strategies and adaptability in the case of future changes to their environments.
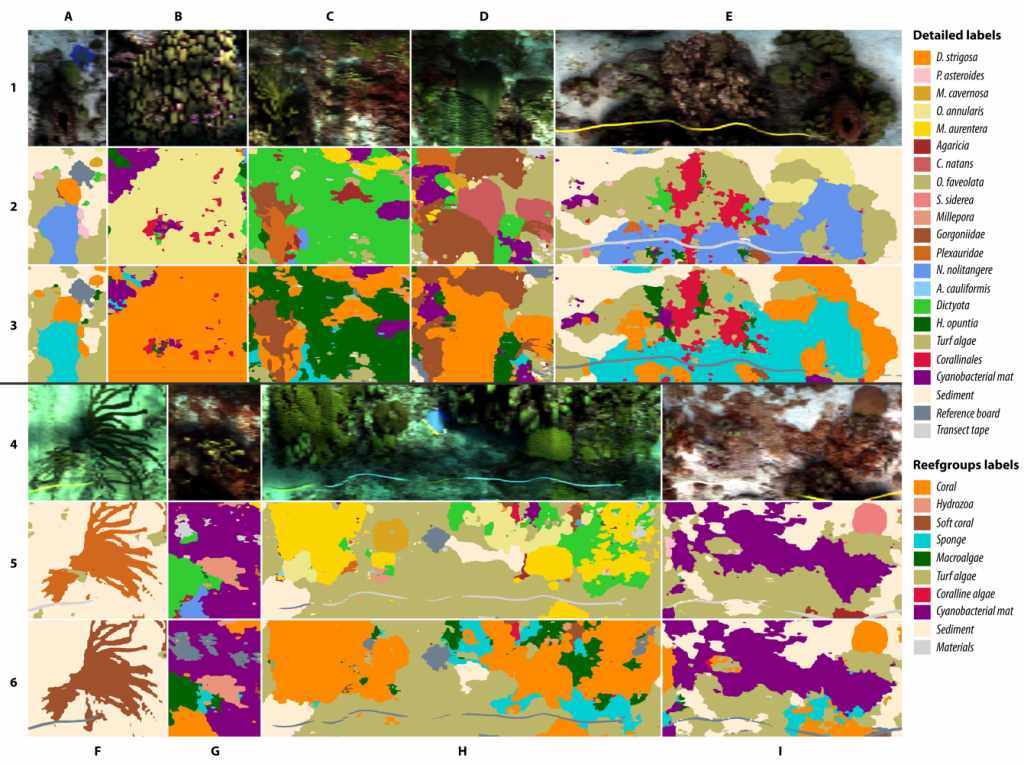 Examples of habitat maps created from data acquired using the HyperDiver camera system in Curacaoan coral reefs in the Caribbean. Habitat maps such as these help scientists elucidate the spatial distributions of organisms and substrate types in their ecosystems. If created over a temporal period they also help understand interactions between the organisms and their environment. Overall, maps are a great monitoring tool that can influence important conservation and ecological management decisions.
Examples of habitat maps created from data acquired using the HyperDiver camera system in Curacaoan coral reefs in the Caribbean. Habitat maps such as these help scientists elucidate the spatial distributions of organisms and substrate types in their ecosystems. If created over a temporal period they also help understand interactions between the organisms and their environment. Overall, maps are a great monitoring tool that can influence important conservation and ecological management decisions.
All of our projects require the base knowledge that comes from answering 2 questions: what class of organisms and substrates can be found in the reefs of Spermonde? And how much/many of each class can be found in the reefs?
To answer these questions we organized secondments, where we could exchange our expertise to make the best out of the data we collected in the field work. Thus, we organized a big 6 week workshop, and with help of the Leibniz Centre for Tropical Marine Research (ZMT), the Max-Planck-Institute for Marine Microbiology (MPIMM), the University of Granada (UGR), and the 4D-REEF central organization board in Naturalis Biodiversity Center in Leiden we invited the three Indonesian students to Bremen, Germany. The workshop took place from the 9th of January until the 17th of February 2023.
So many pictures, so little time!
The data and tools each of us collected on our field trip were very similar, only with differing strategies. We all collected top-down images of the seafloor and wanted to annotate (label) all the organisms we found in them. This is not as trivial as it may sound though. Between Estra and Dino and their team, they collected almost 5000 pictures! Daniel also collected over 60000 images (it’s not a competition, but who’s counting?). No life of ours could be spent annotating all 65000 images. At this point we decided that some automation, inspired by the current AI boom, was needed. But even the smartest of computers needs some manual help from us primitive humans, to learn what exactly to detect in the images.
Automatic point annotations
For the first set of images (~5000) we wanted to annotate a random selection of 50 points (pixels) with a meaningful label. Estra and her students carefully detailed a set of 73 labels (a labelset) describing all organisms and substrates to be found on the coral reef floor. Later we reduced the labelset to 37 labels, for better performance when automating the process. Then, armed with this labelset, the “Coral Point Count with excel extension (CPCe)” software and full commitment, they annotated 821 images.

Although image annotation is done individually, this workshop fostered team discussions and knowledge sharing, leading to a better image annotation process and benthic identification.
This set of human-annotated images was then the input to a machine learning (ML) web service called CoralNet (link). In this great software offered publicly by the University of California in San Diego (UCSD), a set of pre-annotated images is uploaded, used for training a complex neural network, which in turn is used to predict the annotations on new uploaded images, which do not contain any previous annotations. As part of the ESRs’ secondments we all gathered in Bremen Germany, and went over a 3 week workshop, to learn how to set up the CoralNet project and to establish the benefits of such technologies.
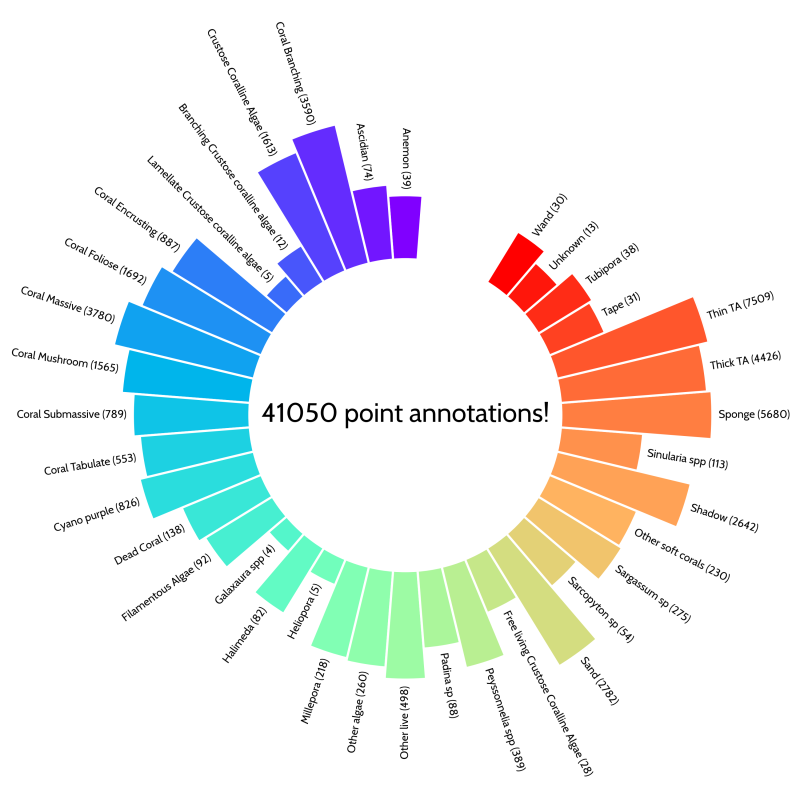
The manual Coral Point Count annotations done by Estra and her students amounted to 41050 points over 37 benthic classes in 821 images! These points were used to train the CoralNet machine learning algorithm.
After training the networks, we tried the first predictions on new images. Because of the complexity of such an ecosystem, the different lighting conditions and possible human error in the annotations, the accuracy of the network was good but not great (57% accuracy). Nonetheless, the CoralNet platform allows to then manually confirm the annotations, by suggesting possible candidates for each new point in each new image. This can reduce the time of annotating significantly (still to be quantified). For comparison, it took three students almost three months to annotate 821 images, whereas CoralNet finished over 4000 images in a couple of hours. There are other parameters that can as well be tweaked to make the predictions more confident. Overall the outcome of the process was positive and we will continue to use its outputs for our study results.
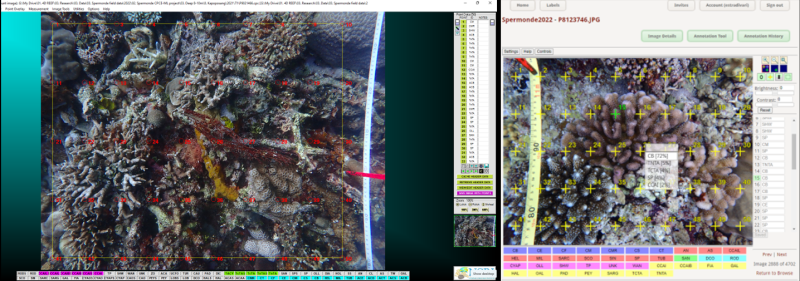
Image with CPCe annotations (left) and a view of CoralNet annotation suggestions with a confidence percentage provided by the AI (right).
Manual polygon annotations
The other big batch of images (~60k) was captured using the HyperDiver (see Daniel’s blog (link) ) by a team of great divers (Dino, Agung, Agus, Dedi, Supardi, Asriadi and Masdar) and one lone snorkeler (Daniel). In contrast to our previous set of images, these were stitched together to create a larger view of the seafloor (an orthomosaic), by a process called photogrammetry. The advantage of these larger scenes is that it is easy to georeference the data, and to create a better spatial description of the seafloor habitat and organism communities. In total we collected 13 of these large plots across 5 islands.
To manually annotate every organism in these large images would be a cumbersome effort. Thus, we decided to annotate a smaller subset of every organism/substrate type that we found, in order to train machine learning networks to automatically annotate the rest of the area for us. We planned the manual annotation part of the process to take place during the last three weeks of the secondment project.
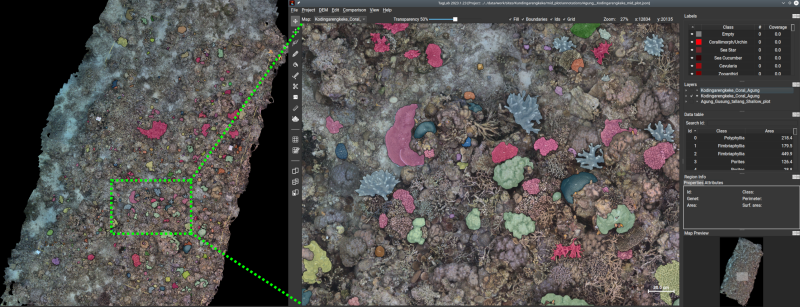
TagLab annotation tool with coral-colony-annotations highlighted. We aimed at annotating as far down the taxonomic tree as possible. In the case of coral, we were able to use genus-level labels, given the high resolution of the orthomosaics and the high level of expertise from our annotators.
By using TagLab (link), a great tool developed by the Visual Computing Laboratory in Italy, we again started annotating many organisms and substrate types in the images, with the only difference being that now we annotated polygons instead of single points. This process is usually tedious and time consuming when using a non-specialized tool, such as a GIS platform, but with the tools that TagLab offers, the process was significantly improved. After a quick introduction to the tools by Daniel, and in only three weeks, the amazing students Phita, Gunawan and Agung, drew and annotated almost 1600 polygons across 3 of the large plots! The labelset used in these annotations has great detail, and covers 36 coral genera, many soft coral genera, sponges, anemones, cyanobacterial mats, turf algae, amongst many other organisms and substrate types. In a further research study these annotations will be used to train an advanced machine learning algorithm to detect individual organisms as well as their surrounding environments with great accuracy. The fully annotated maps created by the AI will then be used to elucidate spatial characteristics of the coral reefs and hopefully set a baseline for future studies.
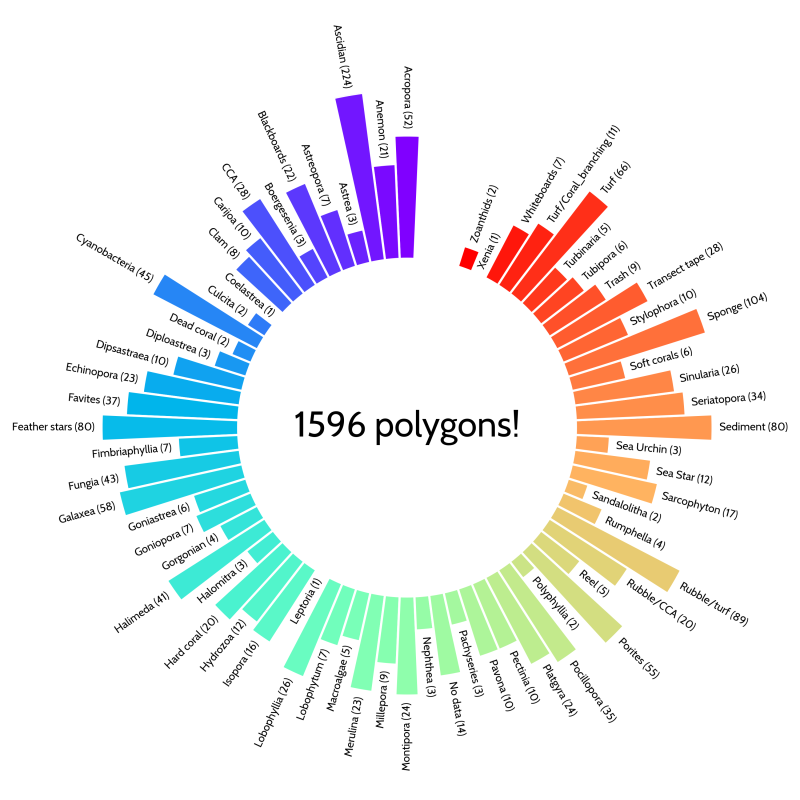
The result of the orthomosaic annotation was great for only 3 weeks of intense annotations. We created 1596 polygons down to genus level for corals, soft corals and macroalgae, as well as other biota and substrate types.
Closing thoughts
We had a great time during the secondment and annotation workshop, which was the conclusion to getting to know each other during the field trip and planning and executing a collaboration with a transdisciplinary and multi-cultural team. We produced knowledge that will help each of us through our career paths and learned to collaborate and communicate with local partners, a much needed skill if we want to make science accessible and visible to researchers worldwide.

Having a delicious Peruvian meal (at the “Pachamama” restaurant in Bremen) with the team and Katherine Maxwell, another 4D-Reef member based in Bremen, was a great way to end the secondment.
Finally, we, the ESRs and UNHAS students, would like to thank everybody who made this small collaborative effort possible, starting by our institutes, the Naturalis team, our fellow ESRs, our supervisors, but specially to the amazing team in Indonesia, who welcomed us from the beginning and created such a nice atmosphere to work in, besides giving valuable input/guidance/sweat-and-hard-work for our projects; Terima kasih!









